Research Overview
Our work focuses on understanding how intracellular bacterial pathogens manipulate host cellular processes to establish infection and evade immune defenses. Specifically, we study two significant human pathogens: Chlamydia trachomatis and Orientia tsutsugamushi. C. trachomatis is an obligate intracellular Gram-negative bacterium that is a leading cause of preventable blindness and sexually transmitted infections, while O. tsutsugamushi is the causative agent of scrub typhus, a severe and sometimes fatal disease endemic to the Asia-Pacific region. While C. trachomatis resides within a membrane-bound vacuole, O. tsutsugamushi replicates in the host cell cytosol. Both pathogens have evolved unique and sophisticated strategies to hijack host signaling pathways, suppress immune defenses, and sustain their intracellular survival.
Using an interdisciplinary approach that combines cell biology, microbiology, genetics, advanced imaging, and proteomics, our lab investigates the cellular and molecular mechanisms these pathogens use to construct and safeguard their intracellular niches. We are particularly interested in studying the type III secreted effector proteins and inclusion membrane proteins (Incs), used by C. trachomatis to maintain its replicative vacuole. We are developing genetic tools to characterize O. tsutsugamushi effectors, with a focus on type I secreted proteins. Our work aims to reveal key mechanisms of pathogenesis and leveraging these insights to better understand host-pathogen interactions to inform therapeutic development.
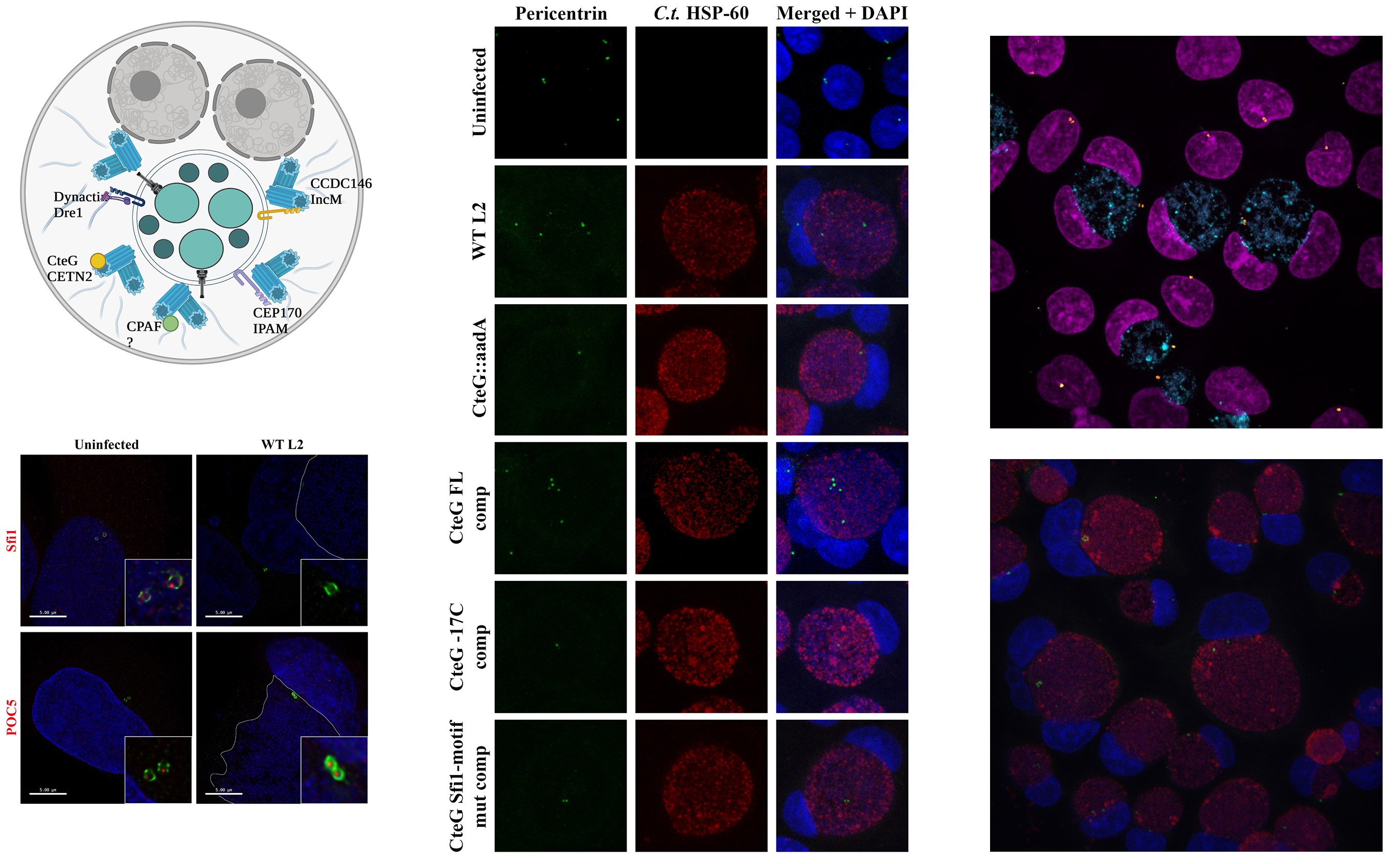
Role of C. trachomatis Effectors in Manipulating the Centrosome and Cell Cycle
C. trachomatis infection has been linked to centrosome amplification, multipolar spindles, and multinucleation—hallmarks of genomic instability observed in cervical and ovarian cancers, where C. trachomatis is a recognized risk factor. Our work focuses on understanding how C. trachomatis effectors, including CteG, drive these cellular abnormalities. We identified CteG as a critical effector that interacts with the centrosomal protein centrin-2 (CETN2), disrupting centrosome duplication pathways. This interaction leads to supernumerary centrosomes, a process we hypothesize enhances infection by altering host cell division and promoting intracellular bacterial survival.
This project also allows us to investigate broader questions about how host cell-cycle machinery and centrosome duplication are manipulated by microbial factors. Leveraging our advanced cellular and molecular tools, we aim to uncover the pathways that govern centrosome amplification and genomic instability during infection. These insights could reveal novel links between infection and oncogenic transformation, shedding light on the underlying mechanisms of infection-associated cancers. By understanding how CteG and other effectors target host cell pathways, we hope to guide strategies that mitigate the cancer risk associated with C. trachomatis infections.
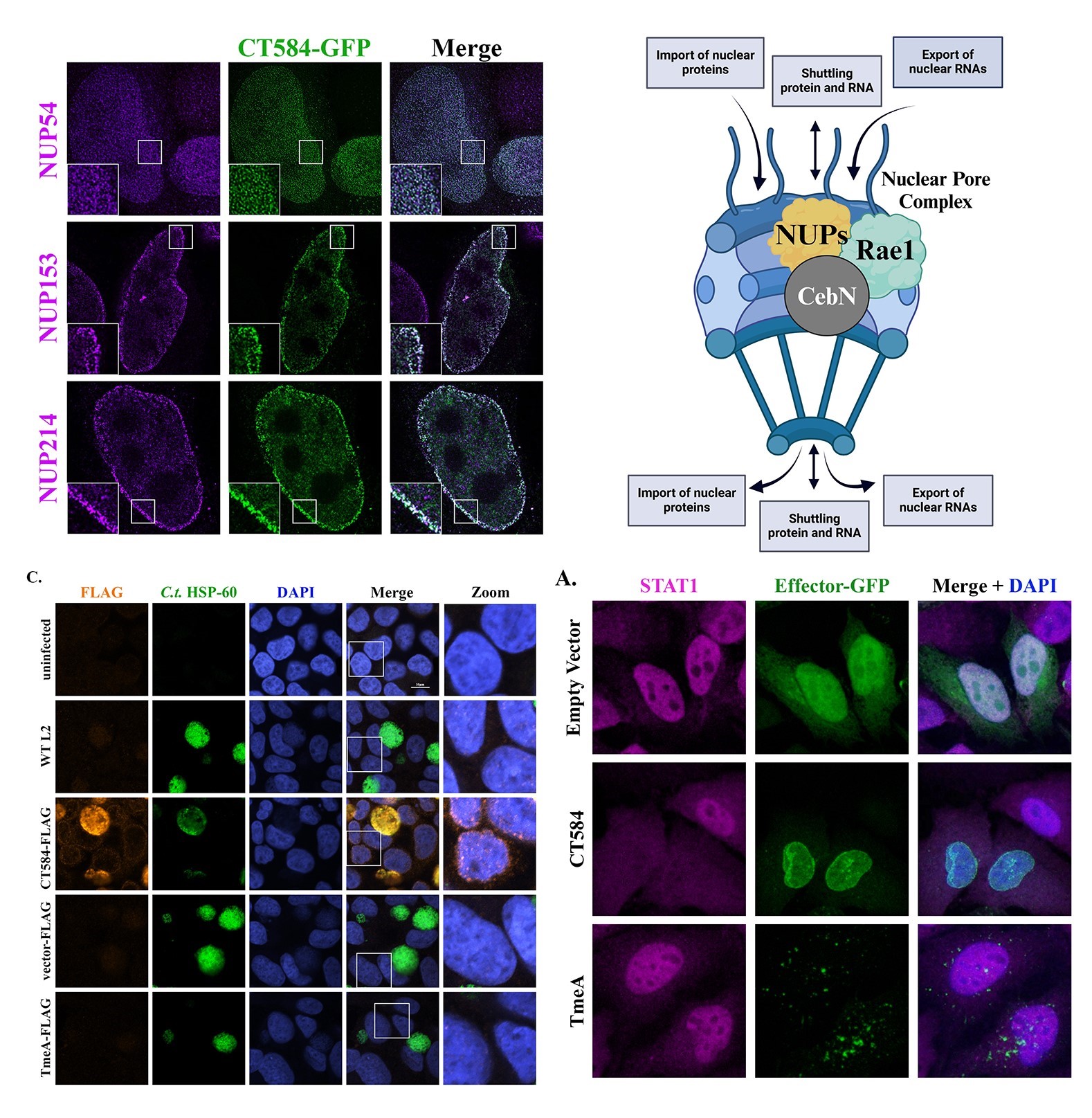
Role of CebN in Disrupting Nucleocytoplasmic Transport and Host Immune Responses
C. trachomatis is adept at evading host immune defenses, establishing persistent infections despite eliciting robust immune responses, including IFN-γ production. Persistent infection can result in significant reproductive health complications, yet the mechanisms by which C. trachomatis achieves persistence remain poorly understood. Our work focuses on CebN, a secreted bacterial effector we identified as the first to target nucleoporins and the mRNA export factor Rae1. These host factors are often hijacked by viruses to disrupt mRNA export, suppress interferon-stimulated gene (ISG) expression, and dampen immune responses, suggesting CebN may function through a similar mechanism.
This project provides an opportunity to explore broader questions about how bacterial effectors manipulate nucleocytoplasmic transport pathways and evade immune detection. Using advanced proteomic and imaging techniques, we aim to elucidate whether CebN’s interactions with nucleoporins and Rae1 suppress ISG expression in both infected and bystander cells, facilitating immune evasion and infection persistence. Understanding how CebN disrupts nucleocytoplasmic transport and translocates into bystander cells could reveal critical insights into bacterial strategies for immune modulation. Ultimately, these findings could inform therapeutic interventions aimed at reducing the burden of persistent C. trachomatis infections.
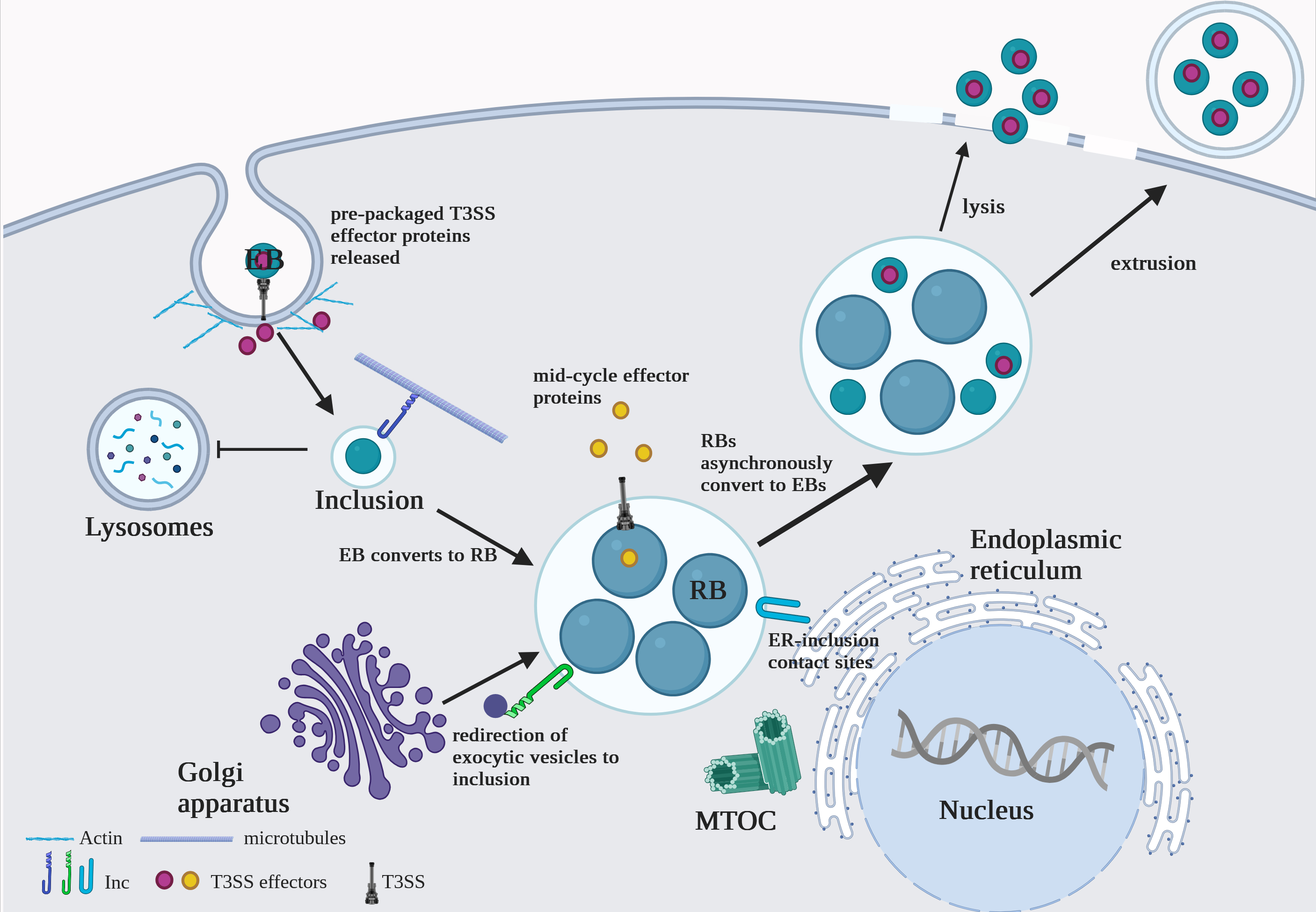
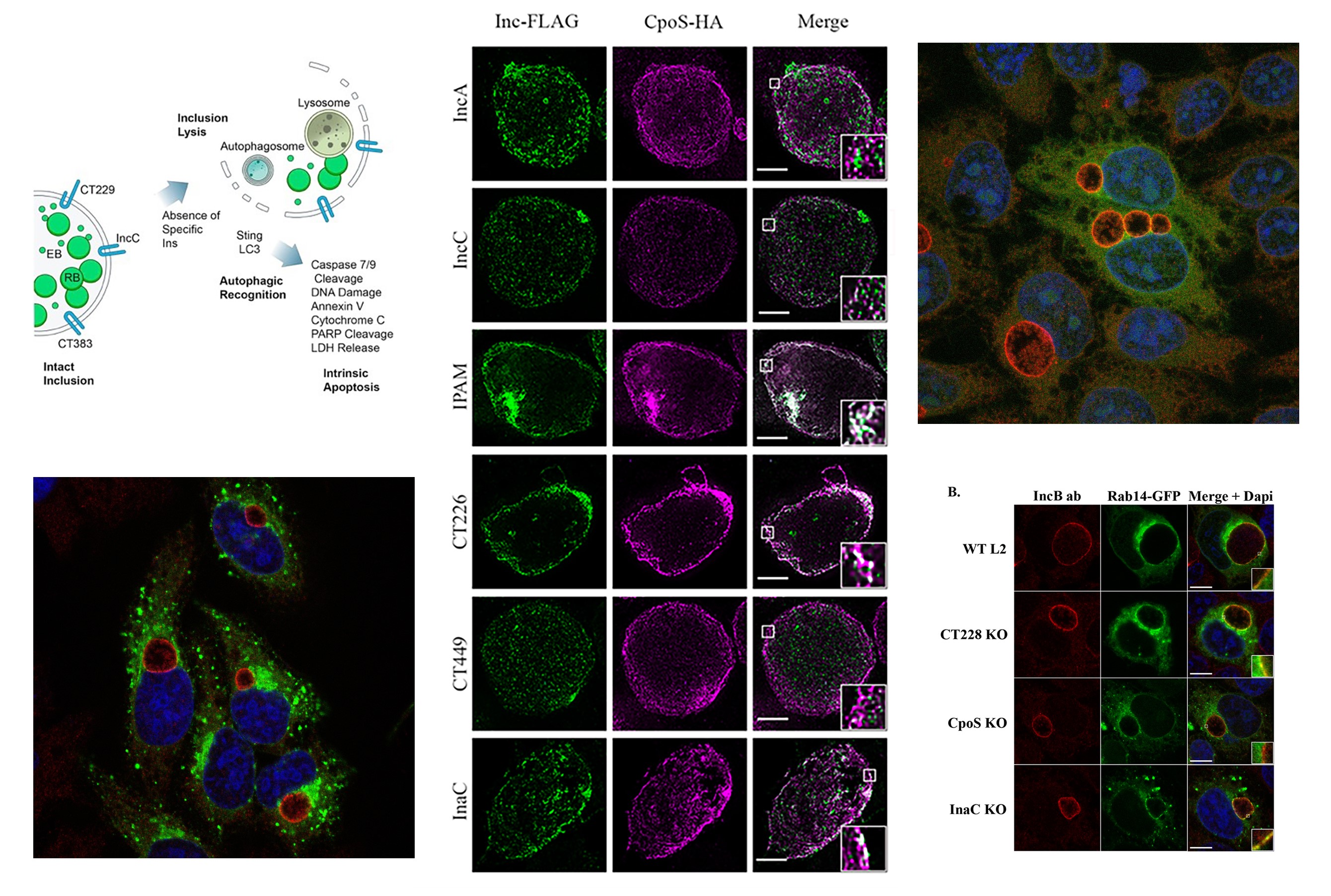
Role of Inc Proteins in Constructing and Protecting the Chlamydia Inclusion
Chlamydia trachomatis relies on a host-derived vacuole, the inclusion, to replicate and evade immune defenses. The inclusion is extensively modified by over 50 type III secreted inclusion membrane proteins (Incs), which play critical roles in maintaining its integrity and shielding it from host degradative compartments. Among these, IncC and CpoS/CT229 are essential for inclusion stability, preventing premature lysis and host cell death. Our work demonstrates that IncC interacts with host factors, such as IFITM3, to inhibit lysosomal targeting, while CpoS forms complexes with other Incs and Rab GTPases to organize the inclusion membrane and facilitate nutrient acquisition.
This project explores fundamental questions about how C. trachomatis orchestrates vesicular trafficking and manipulates host defenses to sustain its intracellular niche. Leveraging advanced biochemical and imaging techniques, we aim to elucidate how IncC and CpoS mediate inclusion stability through interactions with host and bacterial partners. By integrating structural studies with functional assays, this research seeks to reveal novel mechanisms of pathogen adaptation and evasion. Ultimately, these insights will deepen our understanding of C. trachomatis pathogenesis and inform the development of targeted therapeutic interventions.
Development of Genetic Tools for Manipulating O. tsutsugamushi
O. tsutsugamushi, the causative agent of scrub typhus, is a significant global health concern, yet the molecular mechanisms by which this pathogen manipulates host processes remain poorly understood. To address this critical gap, we are developing a genetic manipulation system to facilitate functional studies of O. tsutsugamushi. Our work focuses on identifying and characterizing secreted bacterial effectors, with an emphasis on tetratricopeptide repeat (TPR) proteins, which we hypothesize may represent a novel class of secreted factors essential for modulating host-pathogen interactions.
This project provides a unique opportunity to investigate broader questions about how obligate intracellular bacteria like O. tsutsugamushi co-opt host cellular machinery to promote survival and replication. By employing cutting-edge genetic, proteomic, and imaging techniques, we aim to determine whether TPR proteins contribute to immune evasion or subversion of host signaling pathways. Understanding the roles of TPR proteins and other secreted factors in O. tsutsugamushi pathogenesis will provide critical insights into its virulence mechanisms and how it interacts with its human host. This foundational work could pave the way for the development of novel therapeutic approaches to mitigate the global burden of scrub typhus.